
July 22, 2008
Japan Agency for Marine-Earth Science and Technology
The Archaea World in Marine Subsurface Sediments
Abstract
Drs. Fumio Inagaki and Yuki Morono, Geomicrobiology Group of the Kochi Institute for Core Sample Research (KOCHI), the Japan Agency for Marine-Earth Science and Technology (JAMSTEC; Yasuhiro Kato, President), reported that remarkably abundant Archaea exist in global marine subsurface sediments, as a result of the collaboration between JAMSTEC and the University of Bremen (Prof. Kai-Uwe Hinrichs) in Germany. The population of two domains of life, Archaea and Bacteria, in marine subsurface was studied by intact polar lipids (IPLs) (*2, Fig.2) and DNA extracted from a variety of marine subsurface sediment samples, which included the core sediments collected by the deep-sea drilling vessel “Chikyu” off Japan.
Marine subsurface sediments cover approximately 70% of the Earth’s surface crust. Subsurface sediments was found to be the largest biosphere on Earth because of a large population of Archaea widely distributed in sediments, even in the deep marine subsurface over 300 m below the seafloor. The result contributes to the better understanding of the least explored deep sub-seafloor biosphere as well as their adaptation to the low energy subsurface environments and the evolution of life on our planet.
This will be reported in Nature (online) on July 20.
Lipp, Y. S., Morono, Y., Inagaki, F., and Hinrichs, K.-U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature, in press (doi:10.1038/nature07174).
Background
Marine subsurface sediments harbor remarkably abundant microbial life, more than 1 million cells per cubic centimeter of sediments. Molecular phylogenetic studies revealed that most microbial components are indigenous to marine subsurface, which have not been cultured and characterized yet (Fig.3). Microbiologists and biogeochemists are now challenging to explore the sub-seafloor biosphere with the Integrated Ocean Drilling Program (IODP). In this study, microbial IPLs and DNA were extracted from marine subsurface sediment samples and the population of Bacteria and Archaea was evaluated. The sediments were collected from 16 locations around the world, which involve the core sediment samples obtained by the IODP expeditions and the shakedown cruise of the deep-sea drilling vessel “Chikyu”.
Methods
IPLs were extracted from sediment samples spanning a depth range from 0.01m to 367m below the seafloor. The quantities of bacterial and archaeal IPLs were determined based on the unique structural characteristics for the domain Bacteria and Archaea. The total abundance of biomass-carbon and the correlation between IPL concentration and total organic carbon content were calculated. Also, environmental genomic DNA (*4) was extracted by a new protocol using freeze mill beating in liquid nitrogen (-196°C). Whole genomic DNA was amplified by phi29 polymerase and quantitatively analyzed for bacterial and archaeal 16S rRNA (*5) using quantitative real-time PCR and slot blot hybridization techniques.
Summary of Results
Analysis of IPLs and DNA extracted from global marine subsurface sediments showed that populations of Archaea are notably higher than previously expected. In the sediment buried deeper than 1 m below the seafloor, archaeal IPLs accounted for 87% of the total microbial IPLs (Fig. 4b). Abundance of archaeal 16S rRNA genes was significantly increased up to 40 ~ 50% in average (Fig.5), while the previous estimates were less than a few percent of the total. Given the archaeal IPL and 16S rRNA gene concentrations, it became clear that Archaea significantly contribute to the extant biomass in the marine subsurface.
IPL concentrations showed statistically significant relationship with total organic carbon (TOC) (Fig.6), indicating that the heterotrophic Archaea, which consume buried organic matter as their carbon and energy sources, is the major microbial component in marine subsurface sediments. Based on the IPL concentrations in a wide range of oceanographic settings, the total microbial carbon was calculated to be 90 Pg (petagram: 1015g) in marine subsurface, which corresponds to 0.024 % of TOC (Table 1). This value is much higher than microbial carbon in soil (26 Pg) and aquatic ocean (2.2 Pg).
Future prospectus
It is known that Archaea preferentially inhabit extreme environments such as hot springs, salt fields, deep-sea, and hydrothermal vents. This finding supports the previously argued hypothesis that Archaea well adapt to the extreme and low-nourished environments, such as the deep subsurface, because of their lower cell membrane permeability than bacteria, which lowers maintenance energy by preventing the substrate diffusion and infiltration.
The deep sub-seafloor biosphere is very different from our surface world where there is no sunlight, and fluxes of energy and nutrient are very limited. The presence of abundant Archaea in the marine subsurface is a significant finding for the better understanding of the evolution of life, adaptation to the subsurface environment, and survival strategy under the low energy flux environment. Using samples collected by “Chikyu” and other IODP platforms, the sub-seafloor microbial life and biosphere will be continuously studied, which is one of the most important scientific subjects in the future IODP.
*1:Archaea
Life forms on the Earth are classified into three domains (*2), which are Eukarya, Bacteria and Archaea (refer to Fig.1). Most microbes belong to either Bacteria or Archaea. These two domains of microbial life, Bacteria and Archaea are distinguishable based on their cell membrane lipid structure.
*2: domain
The minimum taxonomic unit of biological class structure classified based on gene sequences of RNA in ribosome (organelle which synthesize protein). All life forms on the earth fall into three domains of Eukarya, Bacteria and Archaea.
*3: intact polar lipids (IPLs)
IPL is the molecular building block of microbial cell membrane lipid structure, which is typically composed of glyceride combined with hydrophobic hydrocarbon chain and hydrophilic sugar or phosphate as a head polar group.
*4: Environmental genomic DNA
Bulk DNA extracted directly from natural samples, such as sediments, which contains genomes of a variety of living organisms. The culture-independent analysis using the environmental DNA is the most powerful molecular ecological tool to understand diversity of naturally occurring microbial community.
*5: 16S rRNA gene
It is a blueprint (gene) of RNA in ribosome that has a function of genetic translation (protein synthesis reaction). The RNA inside all ribosomes are highly conserved (slow structural evolution); therefore, their gene sequences are used widely for phylogenetic identification of individual species. It is clearly distinguishable between the phylogenetic types of Bacteria and Archaea based on the sequence of 16S rRNA gene.
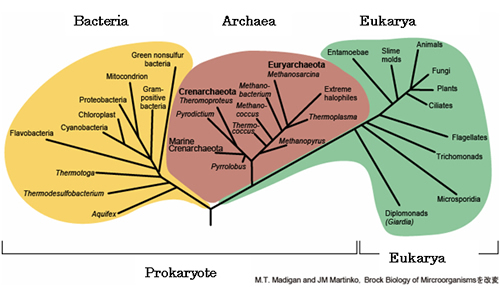
Fig.1. Life on the Earth is comprised of three domains . (*2):Eukarya·Bacteria·Archaea.
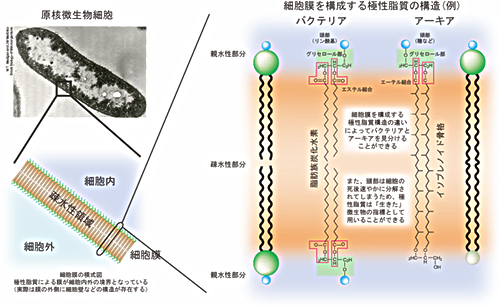
Fig.2. Structure of bacterial and archaeal Intact Polar Lipids (IPLs) in cell membrane. Bacterial IPL has two fatty acid chains connected to glycerol moiety by ester bond. Archaeal IPL structure is composed of isoprenoid chain(s) connected to glycelol moiety by ether bond. IPL has a head group such as sugar and phosphate. IPL is a biomarker for live microbes because the head group structures are decomposed immediately after cell death.

Fig.3. Scanning electron microscopic image of spherical microbes observed in the subsurface sediment obtained by “Chikyu” offshore the Shimokita Peninsula of Japan.
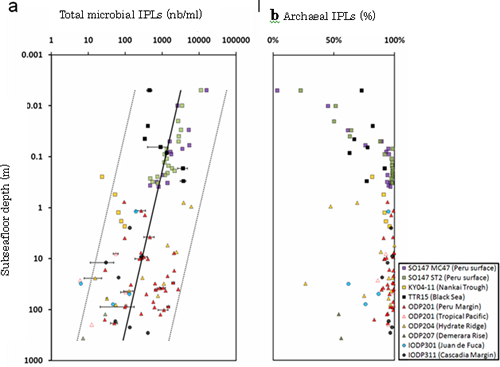
Fig.4.Depth profiles of IPLs in marine subsurface sediments. (a) Concentration of the total IPLs extracted from marine subsurface sediments down to 367 m below the seafloor. (b) Relative ratio of archaeal IPLs to total microbial IPLs.
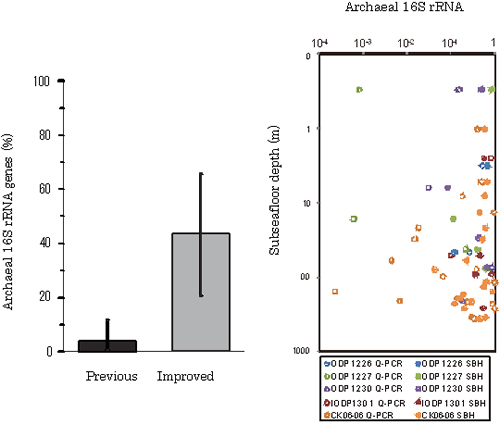
Fig.5.Relative ratio of archaeal 16S rRNA genes. (a) Comparison of archaeal 16S rRNA gene ratios evaluated by the previous and improved methods. (b) Depth profile of relative abundance of archaeal 16S rRNA genes to total microbial 16S rRNA genes.
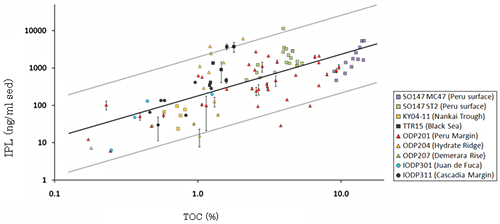
Fig.6.Correlation between IPL concentrations and TOC in marine sediments.
Table 1. Total carbon budget of microbial biomass. Total microbial carbon was estimated to share 0.024 % of TOC.
| Oceanic sediment | Coastal sediment | World marine sediment | |
|---|---|---|---|
| TOC (Pg) | 2.0×105 | 1.7×105 | 3.7×105 |
| IPLs (Pg) | 3.9 | 3.2 | 7.1 |
| Total microbial carbon (Pg) | 50 | 40 | 90 |
Contacts:
- (For the study)
Fumio Inagaki, Group Leader
Yuki Morono, Research Scientist -
Geomicrobiology Group, Kochi Institute for Core Sample Research
The Japan Agency for Marine-Earth Science and Technology - Nobuo Naraki, Manager
-
General Affairs Division, Kochi Institute for Core Sample Research
The Japan Agency for Marine-Earth Science and Technology - (For publications)
Noriyuki Murata, Manager -
Planning Office Press Office
The Japan Agency for Marine-Earth Science and Technology